SDCSB began in 2005 as the San Diego Consortium for Systems Biology to bring together scientists across disciplines and institutions, to promote collaborative research and training, and to support the exchange of ideas and resources.
 In 2010, we became one of the National Centers for Systems Biology funded by the National Institute of General Medical Sciences (NIGMS) (P50 GM085764). Over the past five years our research has led to significant systems biology advances, including genome-wide studies of how stress remodels transcriptional and genetic networks, discovery of an independent metabolic clock coordinating cell growth through cycles of TOR1 activity, demonstration that much of the Gene Ontology can be inferred directly from ‘omics data, prediction of cancer survival time and drug response by an approach called Network Based Stratification and a series of major feats in engineering of synthetic coupled genetic circuits.
In 2010, we became one of the National Centers for Systems Biology funded by the National Institute of General Medical Sciences (NIGMS) (P50 GM085764). Over the past five years our research has led to significant systems biology advances, including genome-wide studies of how stress remodels transcriptional and genetic networks, discovery of an independent metabolic clock coordinating cell growth through cycles of TOR1 activity, demonstration that much of the Gene Ontology can be inferred directly from ‘omics data, prediction of cancer survival time and drug response by an approach called Network Based Stratification and a series of major feats in engineering of synthetic coupled genetic circuits.
The theme of our Center – “From Maps to Models” – addresses an important challenge in systems biology: traversing between network maps and mathematical models, two very successful but so far mostly separate biological representations and modes of study. Network maps tend to be global, static, abstract and descriptive, whereas mathematical models tend to be local, dynamic, detailed and predictive. Guided by this overarching theme, four SDCSB research projects seek to develop a general library of maps and models relevant to fundamental cellular and super-cellular processes, including the spatiotemporal architecture of the genome, whole-cell proteome dynamics, cell-cell communication and heterogeneity and environment-genome interactions. These mapping and modeling activities are fueled by technologies advanced across three SDCSB core platforms.
To stay up-to-date about all our events, please add yourself to our mailing list.





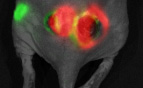


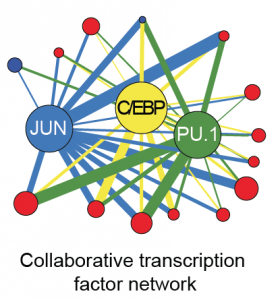 Bing Ren and Chris Glass and colleagues have characterized a large panel of epigenetic features, transcription factor binding patterns and mRNA expression levels among others in macrophages from five strains of mice. By studying the effects of genetic variations, the authors identified a complex network of transcription factors driving differentiation and environmental responses.
Bing Ren and Chris Glass and colleagues have characterized a large panel of epigenetic features, transcription factor binding patterns and mRNA expression levels among others in macrophages from five strains of mice. By studying the effects of genetic variations, the authors identified a complex network of transcription factors driving differentiation and environmental responses.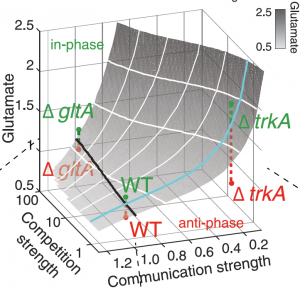 Bacterial biofilms grow in oscillations, with stopping and starting controlled by recently discovered electrical signals. The Suel lab has now found that pairs of bacterial biofilms coordinate their growth behavior depending on environmental conditions. When nutrients are limiting, biofilms oscillate out of phase so that only one is growing at a time. When nutrients though are plentiful, biofilms grow ‘n sync.
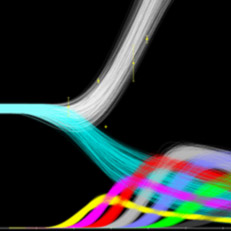
Bacterial biofilms grow in oscillations, with stopping and starting controlled by recently discovered electrical signals. The Suel lab has now found that pairs of bacterial biofilms coordinate their growth behavior depending on environmental conditions. When nutrients are limiting, biofilms oscillate out of phase so that only one is growing at a time. When nutrients though are plentiful, biofilms grow ‘n sync.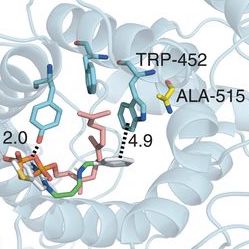 The malaria parasite Plasmodium falciparum can rapidly evolve drug resistance, rendering once life-saving drugs worthless. Elizabeth Winzeler and colleagues recently identified mutations in 83 genes associated with resistance to 37 diverse compounds. These findings suggest new drug targets and advance our understanding of resistance mechanisms.
The malaria parasite Plasmodium falciparum can rapidly evolve drug resistance, rendering once life-saving drugs worthless. Elizabeth Winzeler and colleagues recently identified mutations in 83 genes associated with resistance to 37 diverse compounds. These findings suggest new drug targets and advance our understanding of resistance mechanisms.