Genetics, Bioinformatics and Systems Biology Colloquium
Thursdays, 12:00 pm – 1:00 pm
UC San Diego, Powell-Focht Bioengineering Hall, Fung Auditorium
Complete schedule here
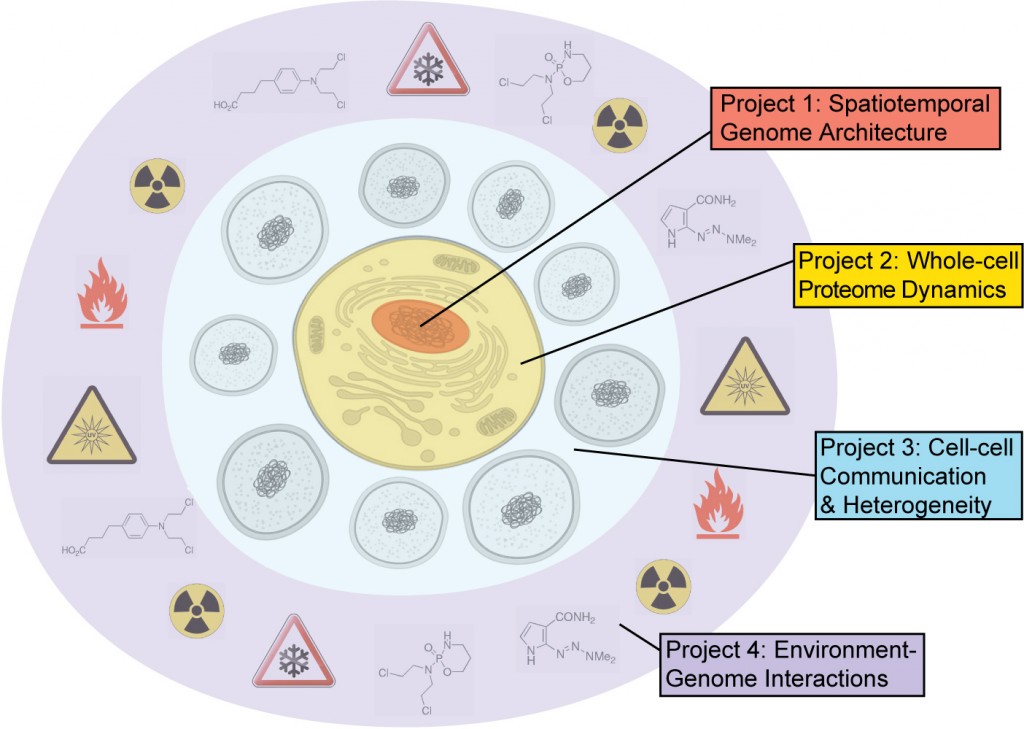
The SDCSB’s four research projects seek to link phenomena at different scales so to gain a unified understanding of both structure (genotype, physical connectivity) and function (phenotype, activities and behaviors, dynamic emergent properties).
Spatiotemporal Architecture of the Genome
A central problem of molecular biology is to understand how transcriptional regulatory elements are selected from the genome so as to specify cellular identity and response. We will use the maps-to-models paradigm to understand how combinations of transcription factors are guided by 3D genome architecture to selectively activate promoters and enhancers. During the mapping phase, ChIP-sequencing, nascent RNA analysis and chromatin conformation capture will define time-dependent changes in transcription factor binding, enhancer activity and 3D genome architecture following exposure of cells to bacteria and viruses. We will test quantitative models for enhancer activity through genome editing and natural genetic variations in mice.
Whole-cell Proteome Dynamics
This project seeks to develop a comprehensive understanding of protein turnover dynamics in response to nutrient availability. A wide variety of systems-wide approaches will be used to characterize key parameters governing protein turnover. Measurements taken from cells grown under a variety of conditions will be used to refine and expand existing mathematical models. By comparing findings across different dividing cells (yeast, mammalian cell lines) and non-dividing cells (neurons), we will identify molecular control principles that govern global rates of protein degradation and synthesis.
Cell-Cell Communication and Heterogeneity
We will develop a quantitative understanding of tissue organization via the maps-to-models paradigm. In the mapping phase we will use quantitative imaging to follow the spatiotemporal dynamics of two model systems: biofilm development and viral infection of epithelia. We will measure with single-cell resolution the spatiotemporal activities of roughly 1 million cells within the context of living tissues. Modeling will involve a reaction-diffusion framework to govern cell-cell communication driven by specific molecular signals. Model predictions will explore the hypothesis that spatiotemporal dynamics of cell communities determine drug resistance and the response to infection.
Environment-Genome Interactions
The genome and molecular circuitry it encodes do not act alone but are embedded in a web of interactions with environmental factors and stimuli such as nutrients, hormones, drugs, toxins and other chemical compounds. We will develop global maps of this environment-genome network using systematic genetic screening coupled to an innovative approach based on genome sequencing of environmentally-resistant evolved isolates. We will model and experimentally validate global functional interactions among regulators of the response and external stimuli including drugs.
In addition, the SDCSB awards a number of seed grants to expand the systems biology community within the existing faculty.
We seek to attract faculty from UCSD, Salk, SBP, Scripps and other local institutes through a highly successful Seed Grant program. This program is aimed at identifying the most exciting new systems biology projects in the San Diego area for inclusion in the SDCSB community. Based on our previous success with this program, the opportunity and reasonable expectation is that some of these seed projects will develop into full research programs within the center or become competitive for funds from other sources.